Synthetic biology to manipulate structure and function in microbial communities
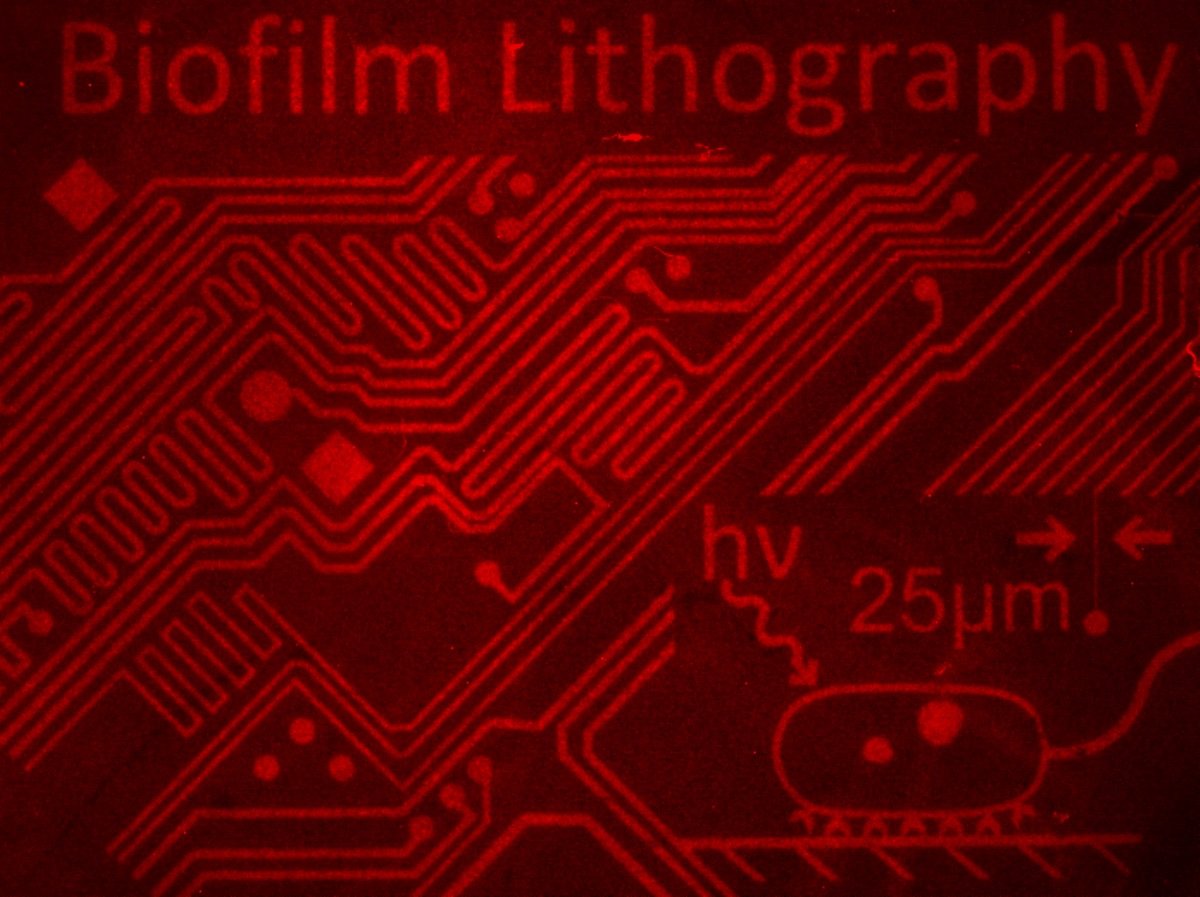
We are interested in using genetic engineering to rationally modulate structure in microbial communities, and learn how these changes ultimately affect community function. For instance, we have developed synthetic biology toolkits that allow for precise patterning of heterogenous biofilm communities, and used these toolkits in conjunction with biophysical modeling to characterize how patterning affects community functions, such as resistance to antimicrobial stress. In ongoing work, we are generalizing these approaches beyond simple communities of laboratory bacterial strains, and into more complex contexts such as the human gut microbiome. These efforts will pave the way toward rational manipulation of microbial communities for desired characteristics, such as the development of next-generation probiotics for modulating gut dysbiosis.
Engineering microbial communities from the ground up

We are applying engineering approaches to generate model microbial communities, with tunable control over community composition and structure. For instance, we have developed in vitro culture platforms with synthetic mucosal surfaces, to profile how surface adhesion modulates function in a defined consortia of human gut microbes. In ongoing work, we are extending these platforms to explore the interplay between community composition, surface adhesion, and a variety of environmental stressors such as antimicrobial exposure or pathogenic invasion. This work will enable precise, quantitative characterization of how host-associated microbial communities critical to human health – such as the human gut microbiome – are affected by changes to their environments.
Metagenomic profiling of community ecology and evolution
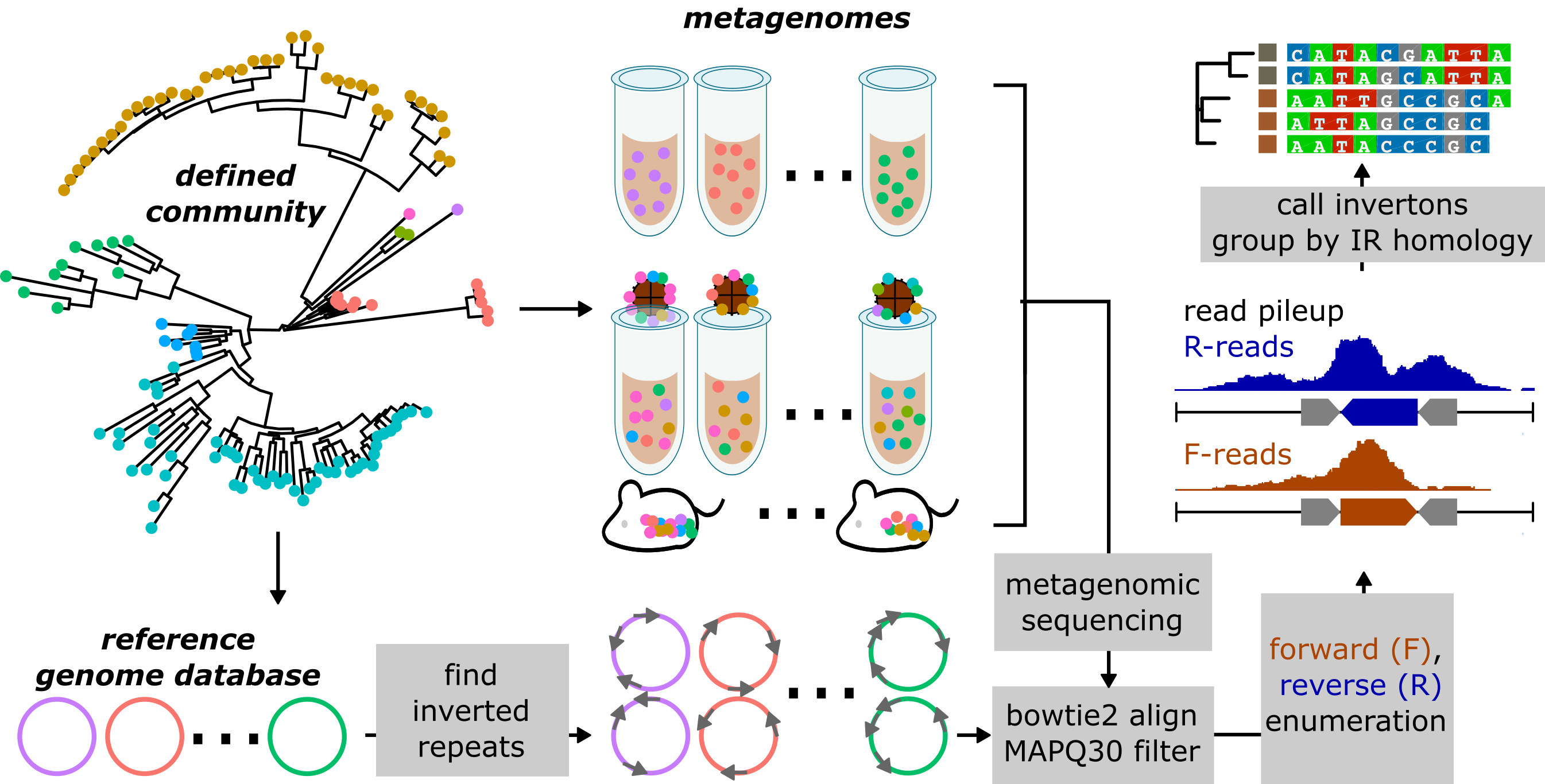
We are developing computational tools to track ecology and evolution in defined microbial communities. These workflows analyze next generation sequencing datasets produced by shotgun metagenomic analysis of microbial communities, to reveal how these communities adapt at an eco-evolutionary level with resolution at the level of individual microbial strains and genes. In ongoing work, we are developing tools to track de novo mutations, genomic inversions, and instances of horizontal gene transfer across bacteria. We envision that such tools will be broadly applicable for a wide range of complex microbial communities, and will allow us to accurately identify critical eco-evolutionary events with consequences for human and environmental health, such as the emergence and spread of antimicrobial resistance.